|
OVERVIEW |
| CSCD (Cancer-specific circRNAs database) is a database developed for cancer-specific circRNAs. CSCD collected available RNA sequencing (total RNA with rRNA depleted or polyA- enriched) datasets from 87 cancer cell line samples. We performed an integrated analysis by using 4 popular algorithms: CIRI, find_circ, circRNA_finder and circexplorer. We then collected circRNAs identified in only cancer samples but not any normal samples. Currently there are 272,152 cancer-specific circRNAs deposited in CSCD. |
|
FEATURES |
| CSCD includes following features which could significantly contribute to circRNA research in cancer: 1) Cancer-specific circRNAs (CS-circRNAs); 2) miRNA target sites in CS-circRNAs; 3) RNA binding proteins (RBPs) binding sites in CS-circRNAs; 4) Potential Open Reading Frames (ORFs) in CS-circRNAs; and 5) Cancer-specific alternative splicing associated with CS-circRNAs. |
|
SEARCH INTERFACE |
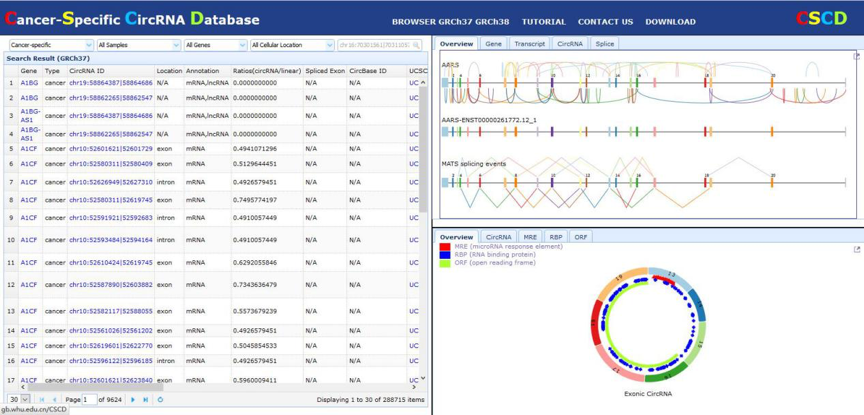
| Main Panel In this panel, users can select or search cancer-specific circRNAs by cell type, gene symbol and cellular location. In the output list, all information of each cancer-specific circRNA are displayed including the host gene, genomic coordinates, location, annotation, ratios (circRNA/linear gene), spliced exon, circbase id, cellular location, prediction algorithm, abundance. Clicking the gene symbol links to the image establishing all circRNAs in all samples. Clicking the each row links this circRNA in special sample. Clicking the UCSC links this circRNA in UCSC genome browser. |
|
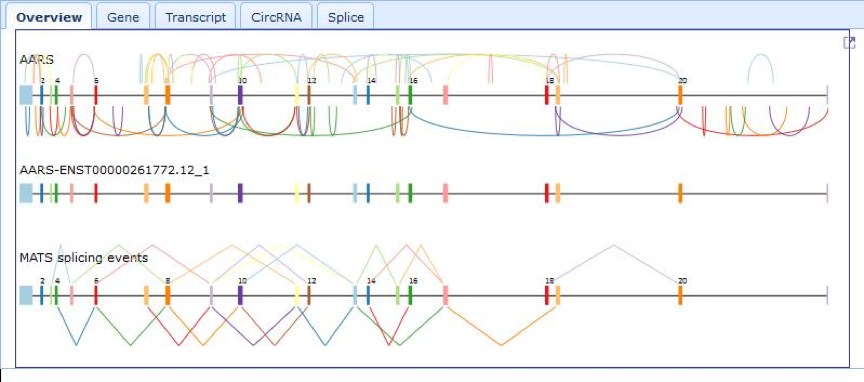
| Right top panel In this panel, users can view the figure of cancer-specific circRNAs. The image displays the gene structures with exons and introns. The donor and accepted site are connected by an arc. Users can view host gene annotation by clicking button 'Gene'. Users can also view the details of cancer-specific circRNA including junction reads, algorithm, location and sample info, by clicking button 'CircRNA'. |
|
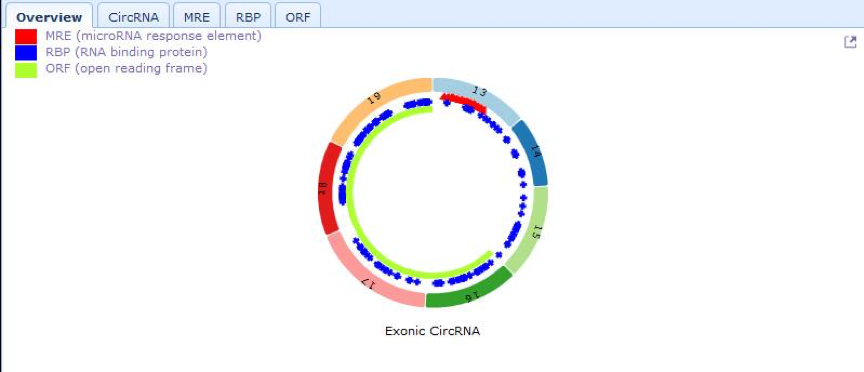
| Right bottom panel This panel displays the image of cancer-specific circRNAs by connecting exons in a circle (different exons are drawing in different colors). In this image, users can view the number and position of MRE (microRNA response element), RBP (RNA binding protein) and ORF (open reading frame) elements located in cancer-specific circRNAs (MRE in red, RBP in blue, ORF in green). By clicking MRE button, users can view the details of MRE site type, start and end position, and microRNA family. By clicking RBP button, users can view the RBP name and coordinates. By Clicking ORF button,users can view the details of ORF position in cancer-specific circRNA. |
|
|
GRCh37 GRCh38 |
| Currently, GRCh37 and GRCh38 were supported.Clicking it to switch two different versions. |
|
DOWNLOAD |
| The complete file of cancer-specific circRNAs including genomic coordinates, gene annotation, number of junction reads are provided in this page. |
|
CITATION |
| Xia S*, Feng J*, Chen K*, Ma Y, Gong J, Cai F, Jin Y, Gao Y, Xia L, Chang H, Wei L, Han L#, He C#. CSCD: A database for cancer-specific circular RNAs. Nucleic Acids Research. 2017. doi: 10.1093/nar/gkx863. |