|
OVERVIEW |
| All the results, including gene annotations, circRNAs, MREs, RBPs, ORFs, full-length sequences, and ASs associated with circRNAs, were organized into a set of interactive MySQL tables. ThinkPHP, an open-source web framework based on a PHP (https://github.com/top-think) and JavaScript library, were used to construct the CSCD 2.0 database. The main function of CSCD 2.0 is composed of three pages: circRNA, miRNA and RBP. All the coordinates in CSCD2 are based on hg38 genome version. |
|
CircRNA page |
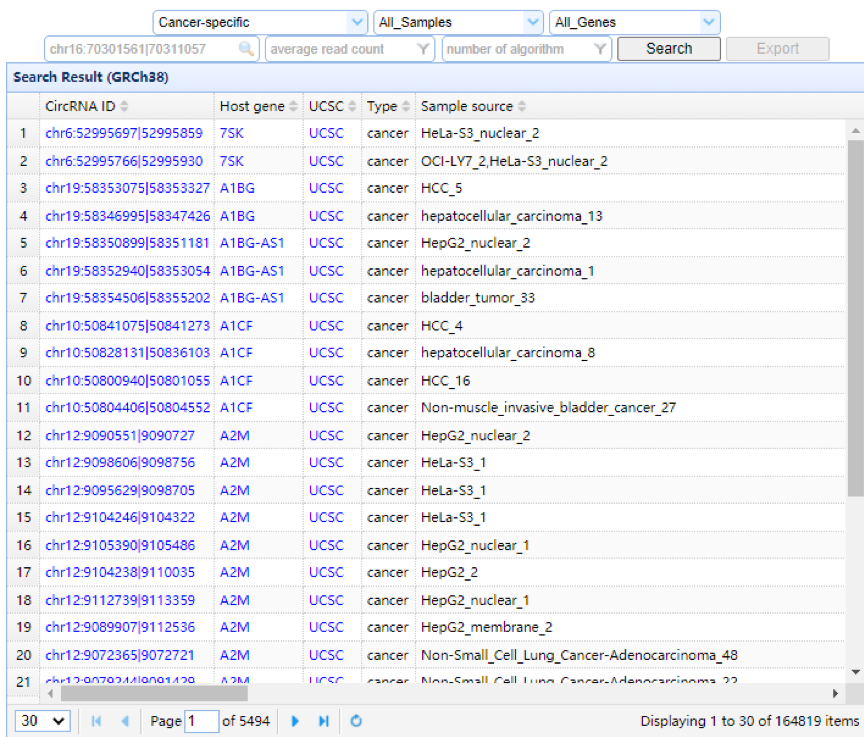
| Left Panel: In this panel, users can browse circRNAs by selecting the sample type, sample name and gene symbol and can search by circRNA ID (e.g., chr10:98425651|98425904), which represents the donor and acceptor sites of each circRNA or gene symbol in the search box. All information, including the circRNA ID, parent gene symbol, UCSC genome browser link, sample type, sample source, constitution, identification algorithm, lncRNA/mRNA annotation, number of junction reads, the average number of junction reads, log2 SRPTM (number of circular reads/number of mapped reads (units in trillion)/read length), average log2 SRPTM, the ratio of circRNA to linear RNA, and circBase ID for each circRNA, is displayed in the table. The filtration and sort function are also provided for users to filter the table by read counts, the number of algorithms, genomic region and sort by columns. The gene symbol links to the upper right panel with all circRNAs across different samples. The circRNA ID links to the lower right panel with a circRNA in a specific sample. |
|
|
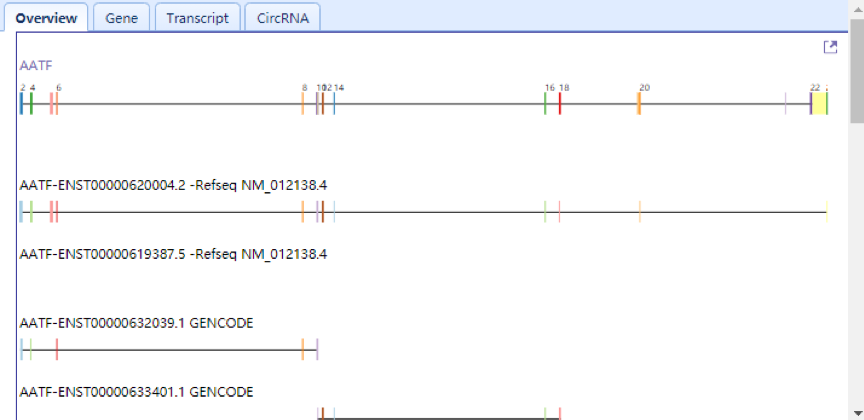
| Upper Right Panel: In this panel, users can view the circRNAs and their linear parent genes in the overview tab. Linear gene structures are displayed with different colored rectangles for exons and black lines for introns, while circRNAs are shown as colorful curves. All transcripts from Refseq and Gencode database of parent gene are also displayed below as annotation. Users can zoom in for a high-resolution image by clicking the top right corner of the panel. All the detailed information is listed in the gene, transcript and circRNA tabs. The circRNA curve links to specific circRNA in the lower right panel. |
|
|
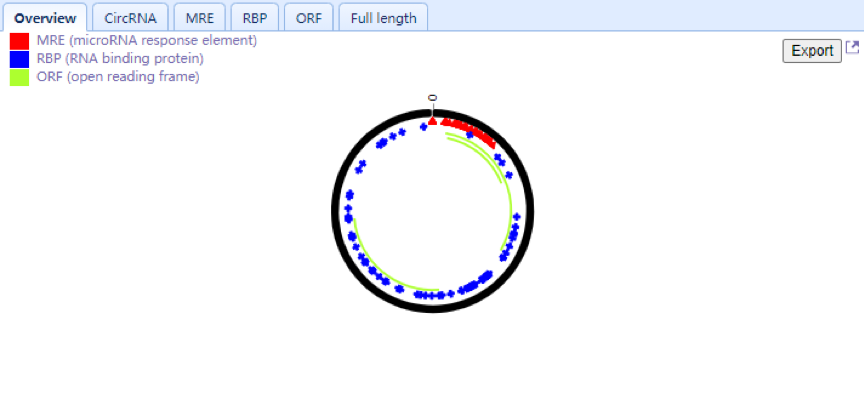
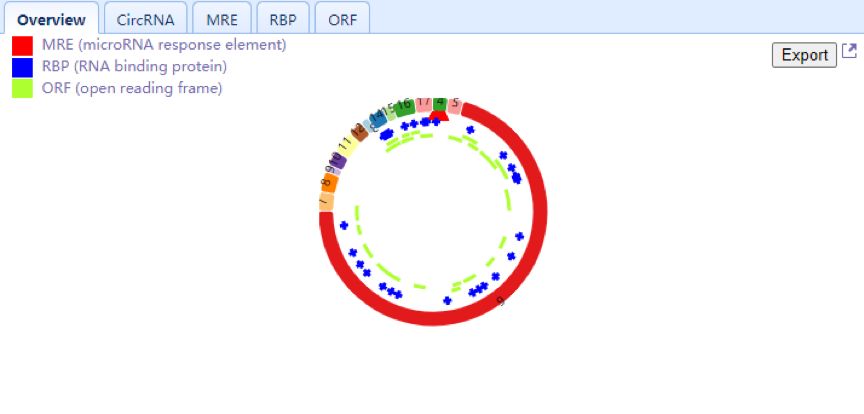
| Lower Right Panel: Users can view selected circRNAs consisting of exons in a colored circle. Each arc with a numeric ID depicts one exon, while introns are displayed in black lines. Users can also view the number and position of the MRE (red triangle), RBP (blue rectangle) and ORF (green arc) elements located in circRNA and check the detailed information through the circRNA, MRE, RBP,ORF and full-length tabs, respectively. An export function is added in the overview Tab allowing users to download the MRE results of selected circRNA. Users can zoom in for a high-resolution image by clicking the top right corner of the panel. |
|
|
|
miRNA page |
| Left Panel: In this panel, users can browse microRNAs by selecting the sample type or microRNA ID (e.g., miR-637) in the search box. Multiple IDs can be checked at the same time. All information, including the circRNA ID, microRNA ID, MSA start (Seed start), MSA end (Seed end), site type, score, energy, Align start (miRNA start), Align end (miRNA end) and algorithm is displayed in the table. The filtration and sort function are also provided for users to filter the table by site type, the number of algorithms, genomic region and sort by columns. CircRNA ID links to the right panel with a circRNA in a specific sample. |
|
|
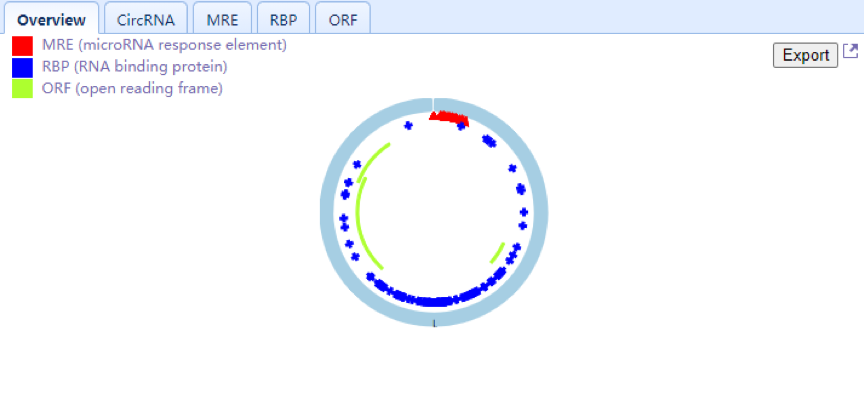
| Upper Right Panel: This panel displays the selected circRNAs that consist of exons in a colored circle. Users can also view the number and position of MREs (red triangle), RBPs (blue rectangle) and ORFs (green arc) located in circRNA and check the detailed information through the circRNA, MRE, RBP and ORF tabs, respectively. An export function is also provided in the overview Tab. |
|
|
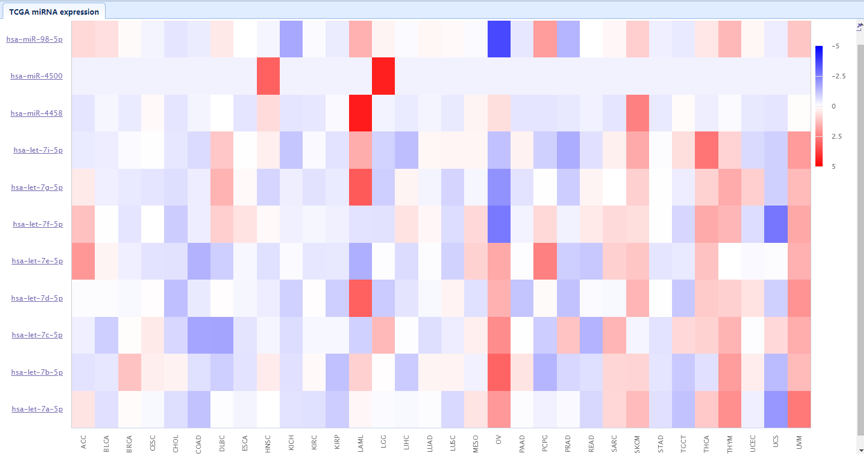
| Lower Right Panel: In this panel, users can view the expression status of selected microRNA in different cancers. The detailed information is listed in the TCGA microRNA expression tab. Expressions in 32 types of cancer are displayed for each microRNA. |
|
|
|
RBP page |
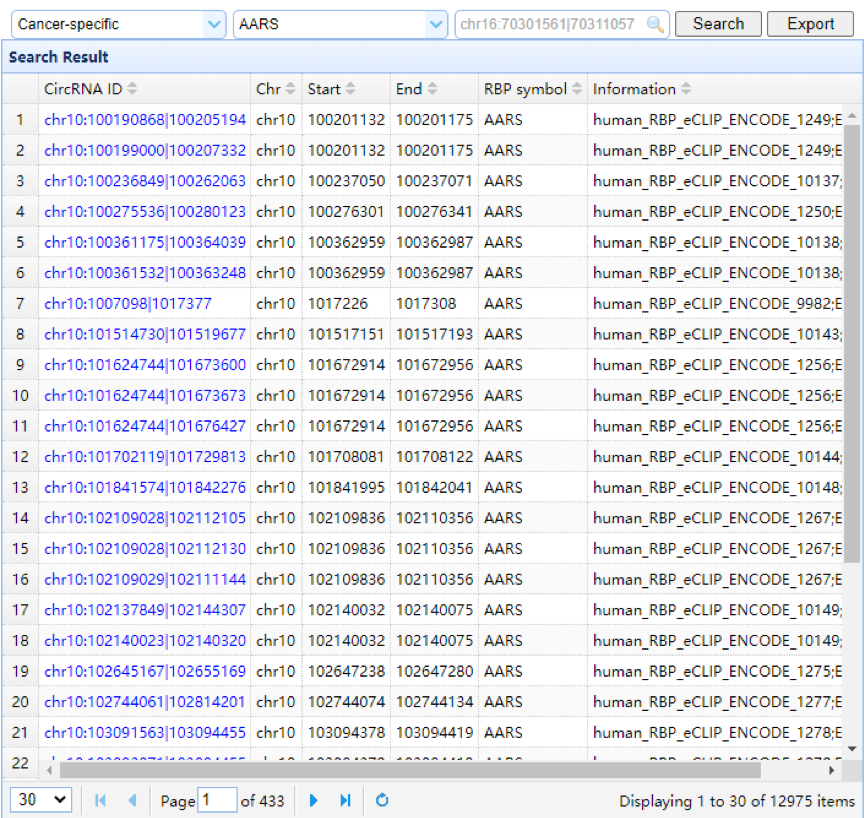
| Left Panel: In this panel, users can browse RBPs by selecting a sample type or RBP gene symbol in the search box. Multiple symbols can be checked at the same time. All information, including the circRNA ID, genomic coordinates of the RBP (chromosome, start, and end), and RBP gene symbol and information, is displayed in the table. Filtration by genomic region and sort by columns are also provided. The circRNA ID links to the upper right panel with a circRNA in a specific sample. |
|
|
| Upper Right Panel: In this panel, users can view selected circRNAs consisting of exons in a colored circle. Users can also view the number and position of MREs (red triangle), RBPs (blue rectangle) and ORFs (green arc) located in the circRNA and view the detailed information through the circRNA, MRE, RBP and ORF tabs, respectively. An export function is also provided in the overview Tab. |
|
|
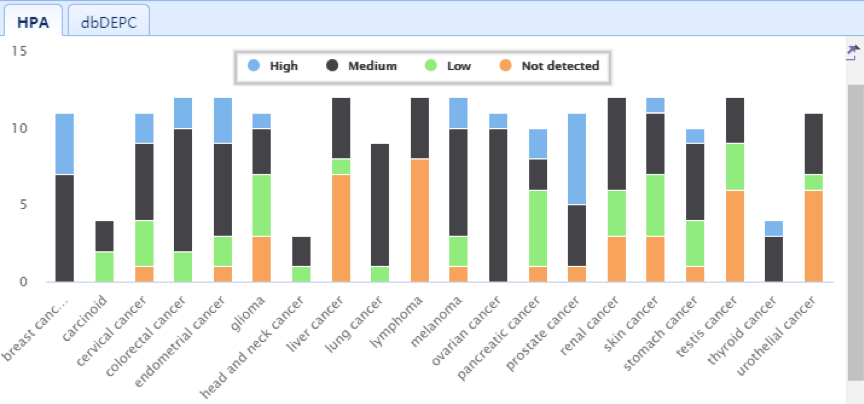
| Lower Right Panel: In this panel, users can view the expression status of selected RBPs in different cancers. The detailed information is listed in the HPA and ORF tabs. Four degrees of IHC staining (high, medium, low, and not detected) are displayed for each RBP. |
|
|
|
DOWNLOAD page |
| All files of circRNAs including genomic coordinates, gene annotation, number of junction reads, ORFs, full-length sequences, and interactions with MREs or RBPs are provided on this page. |